News
Expanse Supercomputer Helps Scientists Develop Promising Model for Studying Flexible Protein Structures
Published August 04, 2024
Zoey Lestyk and Kimberly Mann Bruch, SDSC Communications
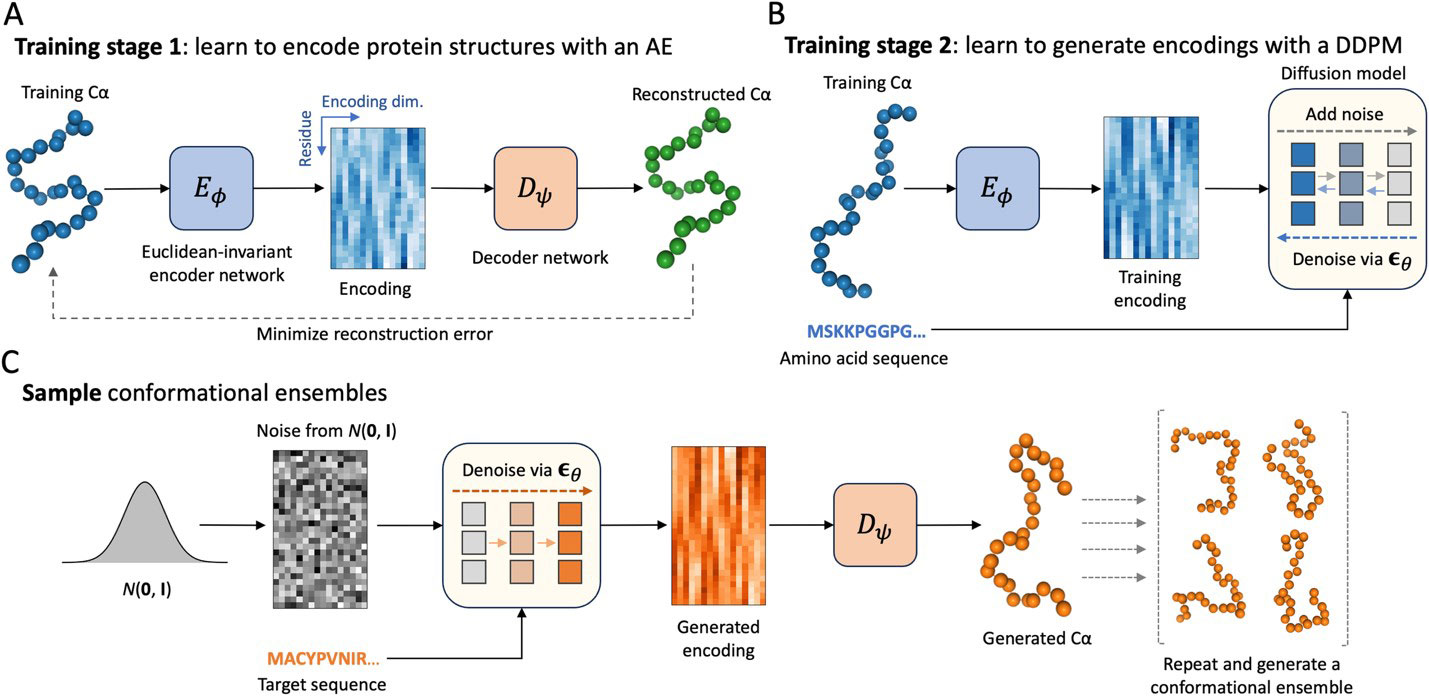
This generative model was created using the Expanse supercomputer via ACCESS allocations. Panels (A) and (B) illustrate the training process of the structural autoencoder model (SAM) while (C) shows how the model generates new samples, starting from random noise and mapping it to 3D structures. Credit: Michigan State University
Using U.S. National Science Foundation (NSF) ACCESS allocations on Expanse at the San Diego Supercomputer Center (SDSC) at UC San Diego, Michigan State University (MSU) researchers recently developed a new model to study flexible protein structures, known as intrinsically disordered proteins (IDPs). The new model might help researchers determine the three-dimensional structure of IDPs for clues about their functionality.
The team published their findings in an article titled Transferable deep generative modeling of intrinsically disordered protein conformations in the PLOS Computation Biology journal. Giacomo Janson and Michael Feig specifically used a diffusion model – a type of machine learning method that can be trained to generate complex objects, such as three-dimensional structures of biomolecules, to conduct this work.
“Our approach called idpSAM (intrinsically disordered protein Structural Autoencoder generative Model) is a diffusion model, a type of deep generative model. These models allow us to generate diverse samples from complex probability distributions, such as the distribution of three-dimensional structures of proteins,” said Feig, who is a biochemistry professor at MSU. “It is really well suited to model the structural ensembles of dynamic proteins like IDPs, and our model is different from existing deep learning approaches that target protein dynamics.”
He said that in idpSAM, an autoencoder first learns a representation of protein geometry and a diffusion model is later trained to sample novel conformations from the encoded space. “This last step allowed us to maintain high computational efficiency,” Feig said, adding that IdpSAM was trained on a large dataset of simulations of disordered protein regions performed with an implicit solvent model called ABSINTH.
“Even though intrinsically disordered proteins have very heterogenous structural ensembles, with idpSAM, a very large, diverse training set may provide enough information to learn general principles with a sufficiently deep network to reach transferability,” explained Janson, an MSU biochemistry research associate. “We believe that deep generative models, such as diffusion models, are a promising method to help us characterize the dynamic behavior of proteins.”
Janson said that current machine learning models used to elucidate three-dimensional ensembles of protein structures are often trained on traditional simulation approaches. However, generative machine learning frameworks using supercomputers like Expanse can be trained in principle to reproduce any target data – including experimental data – and are not limited by the specific functional forms or numerical algorithms that are commonly used in simulations.
“Our work on Expanse with our ACCESS allocations allowed us to collect lots of simulation data and to train large neural network models. This effort showed us how the combination of generative modeling and large training sets can aid us in understanding how dynamical proteins behave,” Janson said. “The finding opens up a whole new way for scientists to explore the complex world of protein dynamics thanks to ACCESS.”
Computational work on Expanse was funded by ACCESS (grant no. BIO230084).

